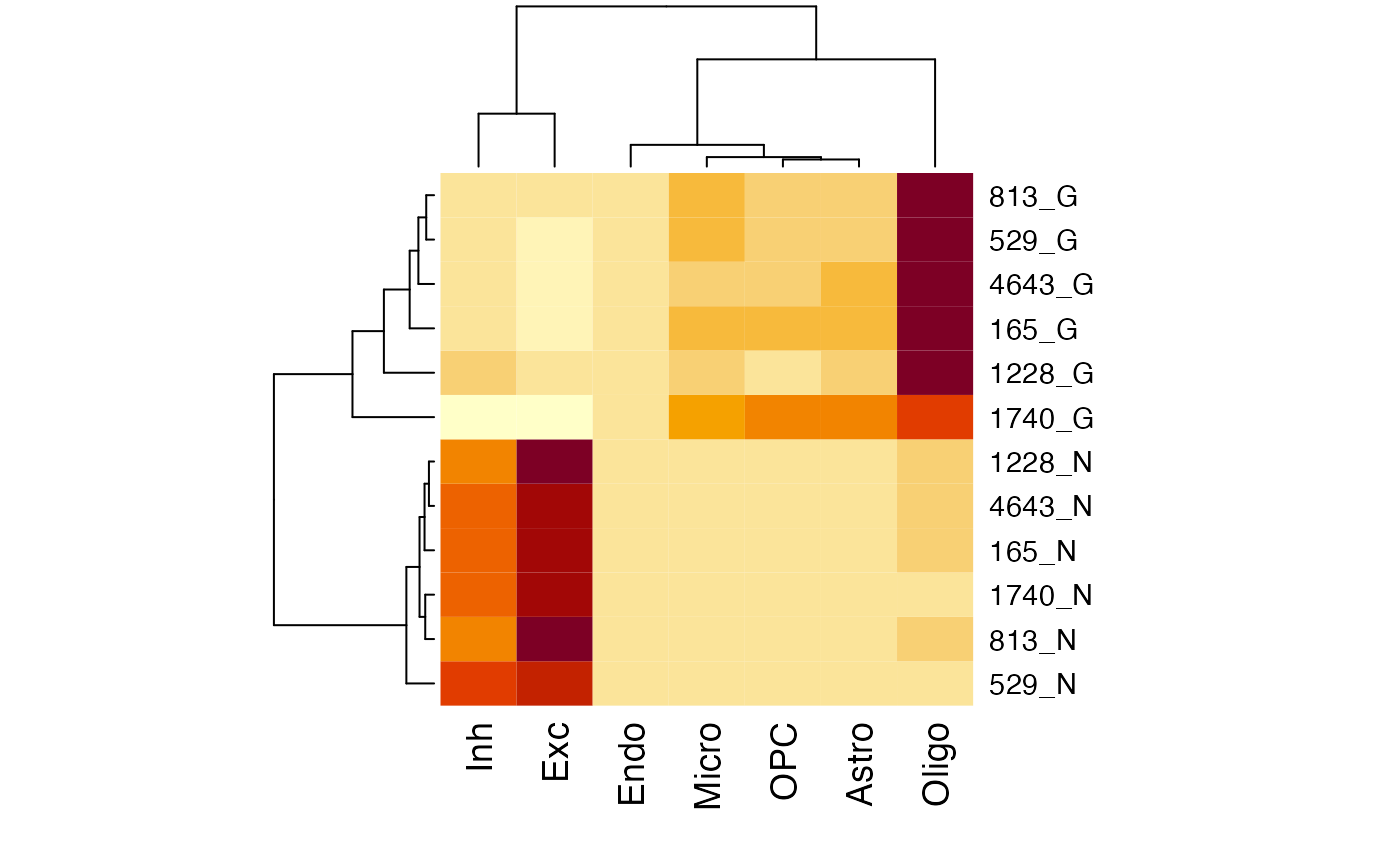Installation
You can install the most recent updates of scMD from github with:
knitr::opts_chunk$set(warning = FALSE, message = FALSE)
# install devtools if necessary
if (!"devtools" %in% rownames(installed.packages())) {
install.packages('devtools')
}
# install the MuSiC package
if (!"scMD" %in% rownames(installed.packages())) {
devtools::install_github('randel/scMD')
}
# load
library(scMD)Data
We’ve made available pre-processed scMD signatures in CSV format for your convenience. You’re welcome to download and use them directly.
If you have your own bulk data for brain tissues and want to apply scMD directly, follow the instructions below to estimate cell type proportions. Please be aware that our estimations focus on seven primary brain cell types: Astrocytes, Endothelial Cells, Excitatory Neurons, Inhibitory Neurons, Microglia, Oligodendrocytes, and OPC.
scMD Example: Guintivano
In this section, we’ll work with a selected subset of the DNAm data from Guintivano et al. as a demonstration. The full Guintivano DNAm dataset is accessible through the Bioconductor package FlowSorted.DLPFC.450k. Our provided subset comprises 12 samples from this data. When utilizing the data, you simply need to specify the technical platform of the bulk data as either “450k_or_850k” or “WGBS”. Additionally, consider setting an output_path to save intermediate results.