Major publications
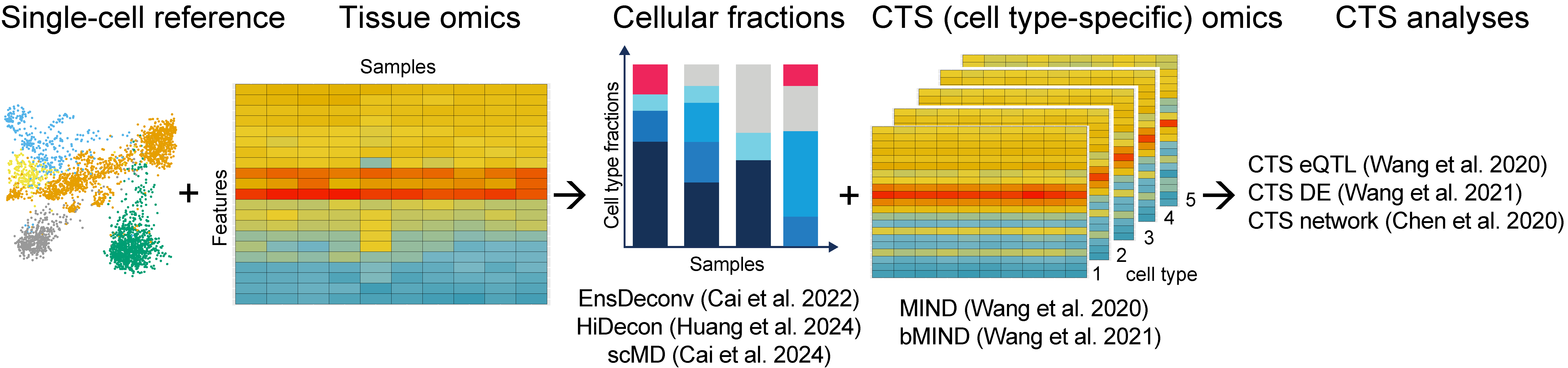
Cellular deconvolution
HiDecon: Huang, Penghui, Cai, Manqi, Xinghua Lu, Chris McKennan, and Wang, Jiebiao+. Accurate estimation of rare cell type fractions from tissue omics data via hierarchical deconvolution. Annals of Applied Statistics, volume 18, pages 1178-1194, 2024. (ICSA Research Poster Award & Student Paper Award Honorable Mention; NESS Student Research Award)
scMD: Cai, Manqi, Jingtian Zhou, Chris McKennan, and Wang, Jiebiao+. scMD facilitates cell type deconvolution using single-cell DNA methylation references. Communications Biology, volume 7, 2024.
EnsDeconv: Cai, Manqi, Molin Yue, Tianmeng Chen, Jinling Liu, Erick Forno, Xinghua Lu, Timothy Billiar, Juan Celedón, Chris McKennan, Wei Chen+, and Wang, Jiebiao+. Robust and accurate estimation of cellular fraction from tissue omics data via ensemble deconvolution. Bioinformatics, volume 38, pages 3004–3010, 2022.
Cell type-specific analyses
bMIND: Wang, Jiebiao+, Kathryn Roeder+, and Bernie Devlin+. Bayesian estimation of cell type–specific gene expression with prior derived from single-cell data. Genome research, volume 31, pages 1807–1818, 2021.
MIND: Wang, Jiebiao, Bernie Devlin, and Kathryn Roeder. Using multiple measurements of tissue to estimate subject-and cell-type-specific gene expression. Bioinformatics, volume 36, pages 782–788, 2020. (Platform talk, ASHG 2018)
Siwei Chen*, Wang, Jiebiao*, Ercument Cicek, Kathryn Roeder, Haiyuan Yu, and Bernie Devlin. De novo missense variants disrupting protein–protein interactions affect risk for autism through gene co-expression and protein networks in neuronal cell types. Molecular autism, volume 11, pages 1–16, 2020.
Statistical genomics
Kyle Satterstrom*, Jack Kosmicki*, Wang, Jiebiao*, Michael S Breen, Silvia De Rubeis, Joon-Yong An, Minshi Peng, Ryan Collins, Jakob Grove, Lambertus Klei, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell, volume 180, pages 568–584, 2020.
mvMISE: Wang, Jiebiao, Pei Wang, Donald Hedeker, and Lin S Chen. Using multivariate mixed-effects selection models for analyzing batch-processed proteomics data with non-ignorable missingness. Biostatistics, volume 20, pages 648–665, 2019.
ofGEM: Wang, Jiebiao*, Qianying Liu*, Brandon Pierce, Dezheng Huo, Olufunmilayo Olopade, Habibul Ahsan, and Lin Chen. A meta-analysis approach with filtering for identifying gene-level gene-environment interactions. Genetic epidemiology, volume 42, pages 434–446, 2018.
MixRF: Wang, Jiebiao, Eric R Gamazon, Brandon L Pierce, Barbara E Stranger, Hae Kyung Im, Robert D Gibbons, Nancy J Cox, Dan L Nicolae, and Lin S Chen. Imputing gene expression in uncollected tissues within and beyond GTEx. The American Journal of Human Genetics, volume 98, pages 697–708, 2016.
* Joint first author; + Corresponding author; student
